Visualize single-cell signals with the signal annotator.
Despite notable efforts in the development of user-friendly software that integrate state-of-the-art solutions to perform single cell analysis, very few are designed for time-lapse data and even less for multimodal problems where cells populations are mixed and can only be separated through the use of multimodal information. Few software solutions provide, to our knowledge, the extraction of response functions from single cell events, such as the dynamic survival of a population directly in the GUI, as coding skills are usually required to do so. We want to study complex data which is often multimodal time lapse microscopy images of interacting cell populations, without loss of generality. With a high need for an easy-to-use, no-coding-skill-required software adapted for images and intended for biologists, we introduce Celldetective, an open-source python-based software with the following highlight features:
- Comprehensive single-cell image analysis : Celldetective ships segmentation, tracking, and measurement modules, as well as event detection from single-cell signals, for up to two populations of interest.
- Integration of state-of-the-art solutions : Celldetective harnesses state-of-the-art segmentation techniques (StarDist, Cellpose) and tracking algorithm (bTrack), as well as the napari viewer where applicable. These algorithms are interfaced to be well integrated and accessible for the target audience, in the context of complex biological applications.
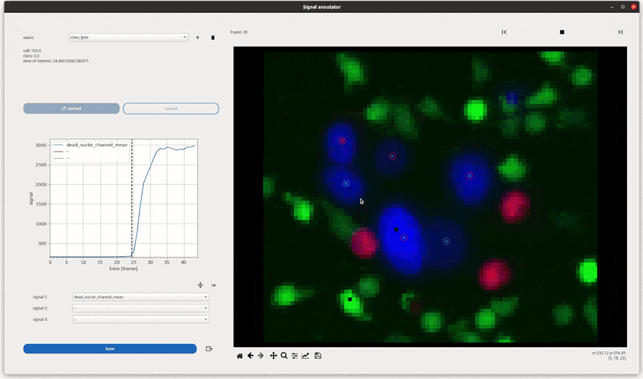
- A framework for event description and annotations : we propose a broad and intuitive framework to annotate and automate the detection of events from single-cell signals through Deep Learning signal classification and regression. The event formulation is directly exploited to define the population’s survival responses.
- A neighborhood scheme to study cell-cell interactions : we introduce a neighborhood scheme to relate the spatio-temporal distribution and measurements of two cell populations, allowing the study of how cell-cell interactions affect single-cell and population responses.
- Deep Learning customization in GUI : Celldetective simplifies the specialization of Deep Learning models or the creation of new ones adapted to user data, by facilitating the creation of training sets and the training of such models, without having to write a single line of code.
- In-software analysis : Celldetective ships visualization tools to collapse single-cell signals with respect to an event, build survival curves, compare measurement distributions across biological conditions.
- A library of segmentation and signal models: we created specific models to investigate a co-culture of MCF-7 cells and primary NK cells, that are available directly in the software with a large collection of generalist models developed by the StarDist and Cellpose teams, which are a perfect starting point to segment single cells in a new biological system.
- Accessible and open source : Celldetective does not require any coding skills. The software, its models and datasets are made fully open source to encourage transparency and reproducibility.
Celldetective: an AI-enhanced image analysis tool for unraveling dynamic cell interactions. R Torro*, B Dı̀az-Bello, D El Arawi, K Dervanova, L Ammer, F Dupuy, P Chames, K Sengupta, L Limozin*. Reviewed preprint, ELIFE (2025) DOI: 10.7554/eLife.105302 https://celldetective.readthedocs.io/en/latest/adcc-example.html

You must be logged in to post a comment.